How DNA sequencing with next generation sequencing (NGS) works
DNA sequencing determines the order of the bases that make up DNA. It provides information about organisms in areas as diverse as population genetics, epidemiology, organism identification, genotyping, rare variant identification, cancer and rare disease research, gene editing confirmation, and gene-environment interactions.
NGS is a high-throughput technology that determines the sequence of a sample all at once by using parallel sequencing. Traditional Sanger sequencing determines the sequence of a sample one section at a time. Sequencing thousands of gene fragments simultaneously with NGS reduces time and cost associated with sequencing and increases the coverage quality and data output.
NGS workflow
Most NGS protocols follow a similar workflow (Figure 1). The first step is nucleic acid extraction, either DNA or RNA. This nucleic acid material must be prepared for sequencing by converting it into libraries. Library preparation involves adding adapters, which allow the samples to be indexed (barcoded and identified).
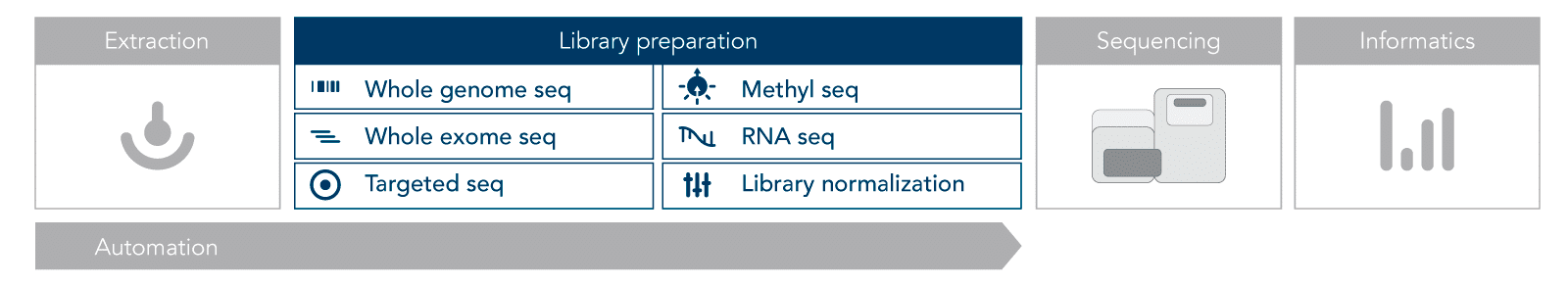
Illumina® sequencing is performed using sequencing-by-synthesis on a flow cell. Short-read sequencing generates 100–300 base pair lengths called reads. The reads go through quality control and are aligned to a reference before being evaluated for significant characteristics.
Learn more about NGS library prep and why it is important in the Why library prep is essential to sequencing DECODED.
What are the types of NGS?
There are many types of NGS for DNA, RNA, DNA-protein interactions, and methylation. The different NGS approaches can be largely thought of as targeted and untargeted.
Untargeted NGS, which examines all the nucleic acids in a sample include approaches like:
While targeted NGS relies on primers or probes to target distinct genetic regions of a sample. Examples of targeted NGS approaches include:
- Targeted RNA sequencing
- Targeted methyl sequencing
- Whole exome sequencing
- Amplicon sequencing
- Anchored Multiplex PCR (AMP™) chemistry
NGS application include genotyping, cancer research, infectious disease research, and genetic diseases research.
Learn more about NGS
To learn more about these approaches, download the Next generation sequencing guide. This extensive application guide provides chapters on NGS workflow, as well as types of sequencing and applications. Data analysis and new sequencing platforms are also discussed. The guide is written by IDT scientists, and is free—learn more.
RUO23-1727_001.1