What are PAM requirements for CRISPR?
In CRISPR experiments, protospacer adjacent motifs (PAM) are short sequences of genomic DNA located directly next to the targeted modification site. PAM sequences are an important requirement for any CRISPR experiment as they are the parts of DNA that the Cas nuclease recognizes. PAM sequences act as a signal for Cas nucleases to let them know they have found the correct modification site.
The Streptococcus pyogenes Cas9 (spCas9) nuclease recognizes an NGG PAM (Figure 1), other nucleases recognize different PAMs. For example, the Cas12 nuclease from Acidaminococcus sp. (asCas12a or Cpf1) relies on a TTTV sequence (Figure 2). Cas nucleases won’t cut the PAM site itself but modify the DNA upstream or downstream of the PAM site. More specifically, Cas nuclease is thought to destabilize the adjacent sequence, allowing interrogation of the sequence by the crRNA, and resulting in RNA-DNA pairing when a matching sequence is present [1,2].
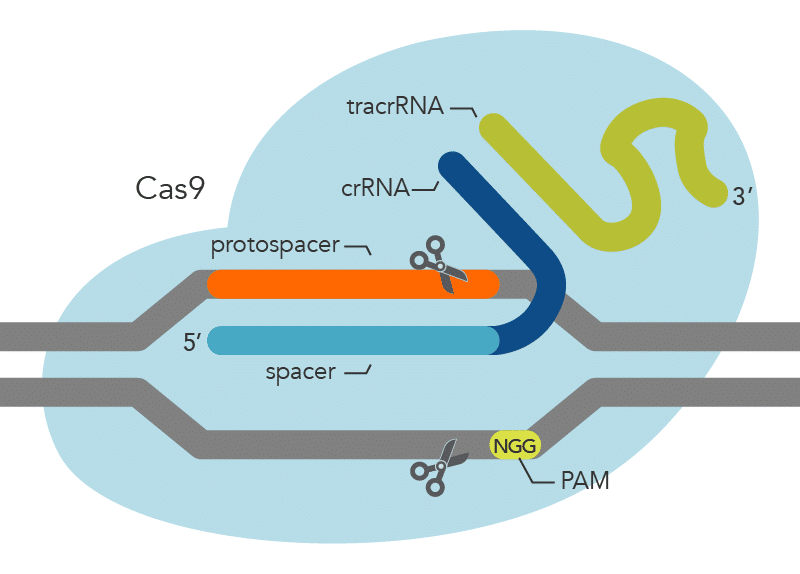
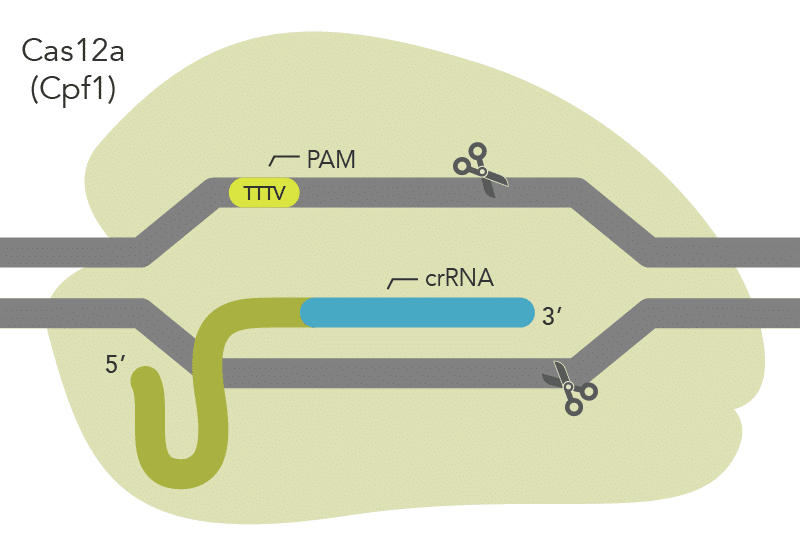
However, the targeted site in the genome may not always be downstream from a NGG or TTTV PAM sequence. Fortunately, in addition to the variety of PAMs offered by wild-type Cas nucleases isolated in different bacteria, researchers have also created modified Cas enzymes to recognize an even larger variety of PAM sequences. This provides greater flexibility when selecting modification target sites in a genome.
Engineering Cas nucleases to recognize novel PAMs
One example of Cas nucleases modified to recognize different PAMs are the Alt-R™ CRISPR-Cas12a nucleases, which come in two formats:
- Alt-R Cas12a V3 which recognizes a TTTV PAM sequence
- Alt-R Cas12a Ultra which works with a TTTN (N = any nucleotide) PAM site
These Cas nucleases are recombinant Cas12a enzymes from Acidminococcus sp. (V3 Cas) and Lachnospiraceae (Ultra Cas). The Alt-R Cas12a Ultra nucleases have higher on-target potency than their wild-type Cas12a nuclease counterparts. These mutants recognize TTTT PAM sites in addition to TTTV motifs, which increases their target range for genome editing studies. Additionally, the Alt-R Cas12a Ultra mutant has an increased temperature tolerance, providing more flexibility for gene editing in systems that require lower culture temperatures.
Other Cas nuclease variants from different bacterial species that recognize different PAM sequences are also exist (Table 1)[3,4].
Table 1. Overview of diverse Cas nucleases and their associated PAM sequence.
| Cas name | PAM sequence (5’—3’) |
|---|---|
| SpCas9 | NGG |
| SaCas9 | NNGRRT |
| CjCas9 | NNNNACAC |
| AsCas12a | TTTN |
| LbCas12a | TTTN |
| AsCas12f1 | NTTR |
| PlmCas12e | TTCN |
Beyond modifying Cas enzymes to recognize different PAMs, researchers have also worked to enhance the specificity of Cas enzymes to reduce off-target effects. Alt-R S.p. HiFi Cas9 nuclease is one example of a Cas9 enzyme that has been modified with this goal in mind. Relative to the wild-type, the Alt-R S.p. HiFi Cas9 nuclease has been shown to dramatically reduce off-target editing effects (Figure 3).
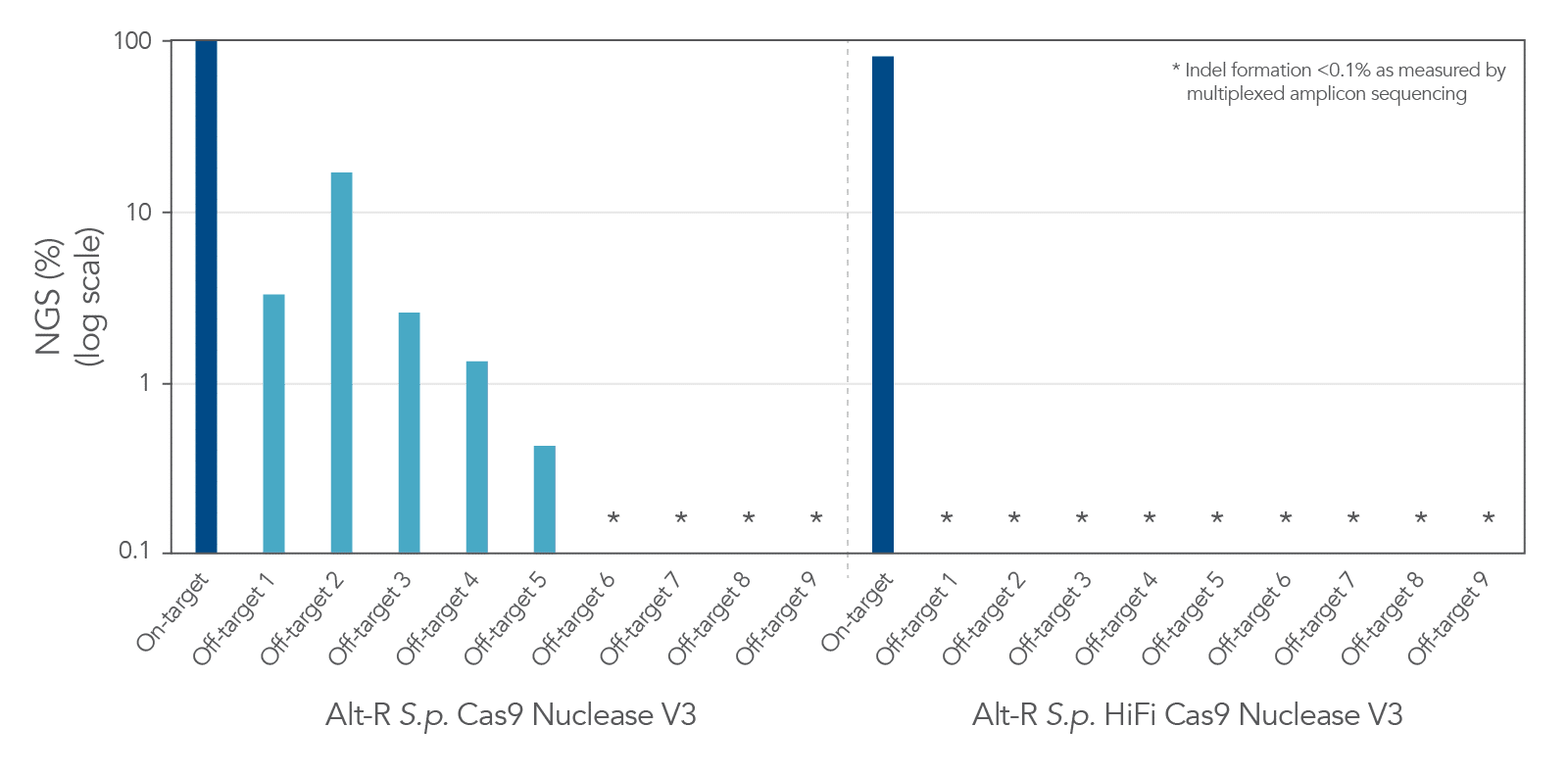
Continuing efforts by researchers to isolate novel Cas nucleases as well as modify already identified Cas enzymes have greatly increased the expanded ways that CRISPR can be applied to gene editing applications.
References
- Anders C, Niewoehner O, Duerst A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569-573.
- Sternberg SH, Redding S, Jinek M, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62-67.
- Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481-485.
- Xin C, Yin J, Yuan S, et al. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat Commun. 2022;13(1):5623.
RUO24-2780_001