Locked nucleic acids improve SNP genotyping and other hybridization assays
Affinity Plus DNA & RNA Oligonucleotides and Affinity Plus qPCR Probes contain locked nucleic acid bases and are ideal for applications requiring increased hybridization affinity or enhanced stability in their oligo designs. The locked nucleic acid bases included in these sequences:
- Increase hybrid melting temperature (Tm), and hybridization affinity
- Enhance sequence stability and nuclease resistance
- Enhance discrimination of thermodynamically similar samples, such as single nucleotide polymorphisms (SNP) and transcript variants
- Facilitate the design of shorter probes, which have better quenching and a higher signal-to-noise ratio than unmodified probes
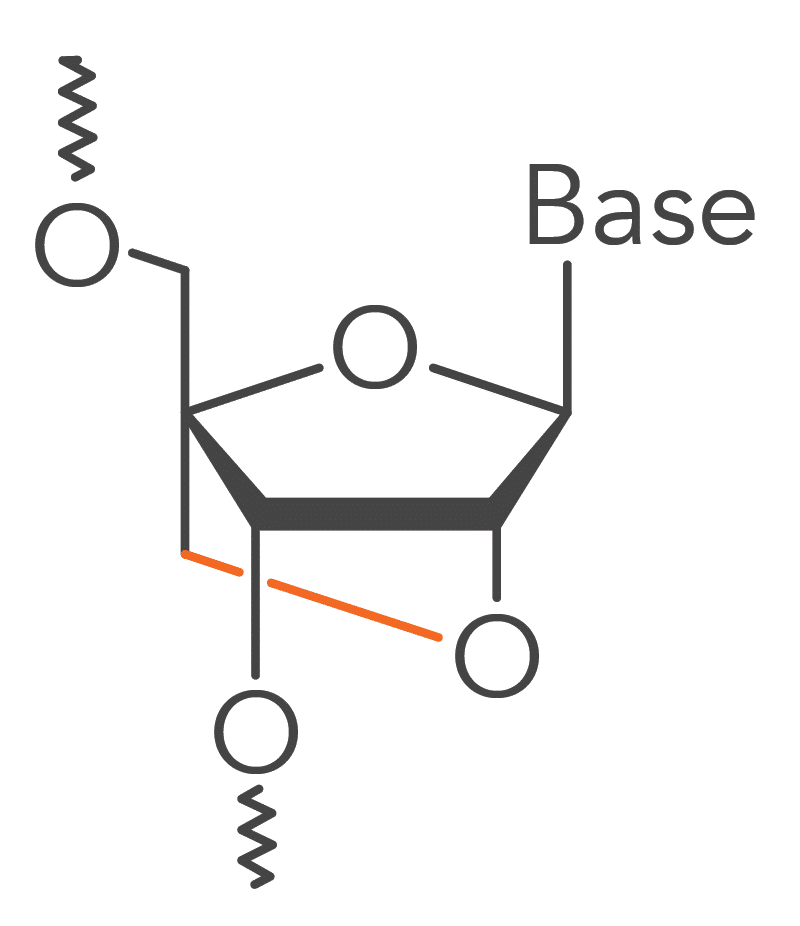
Locked nucleic acid bases impart heightened structural stability to Affinity Plus DNA & RNA Oligos and Affinity Plus qPCR Probes [1]. This enhanced stability is derived from a methylene-bridge bond that links the 2′ oxygen to the 4′ carbon of the RNA pentose ring of this modified RNA base (Figure 1).
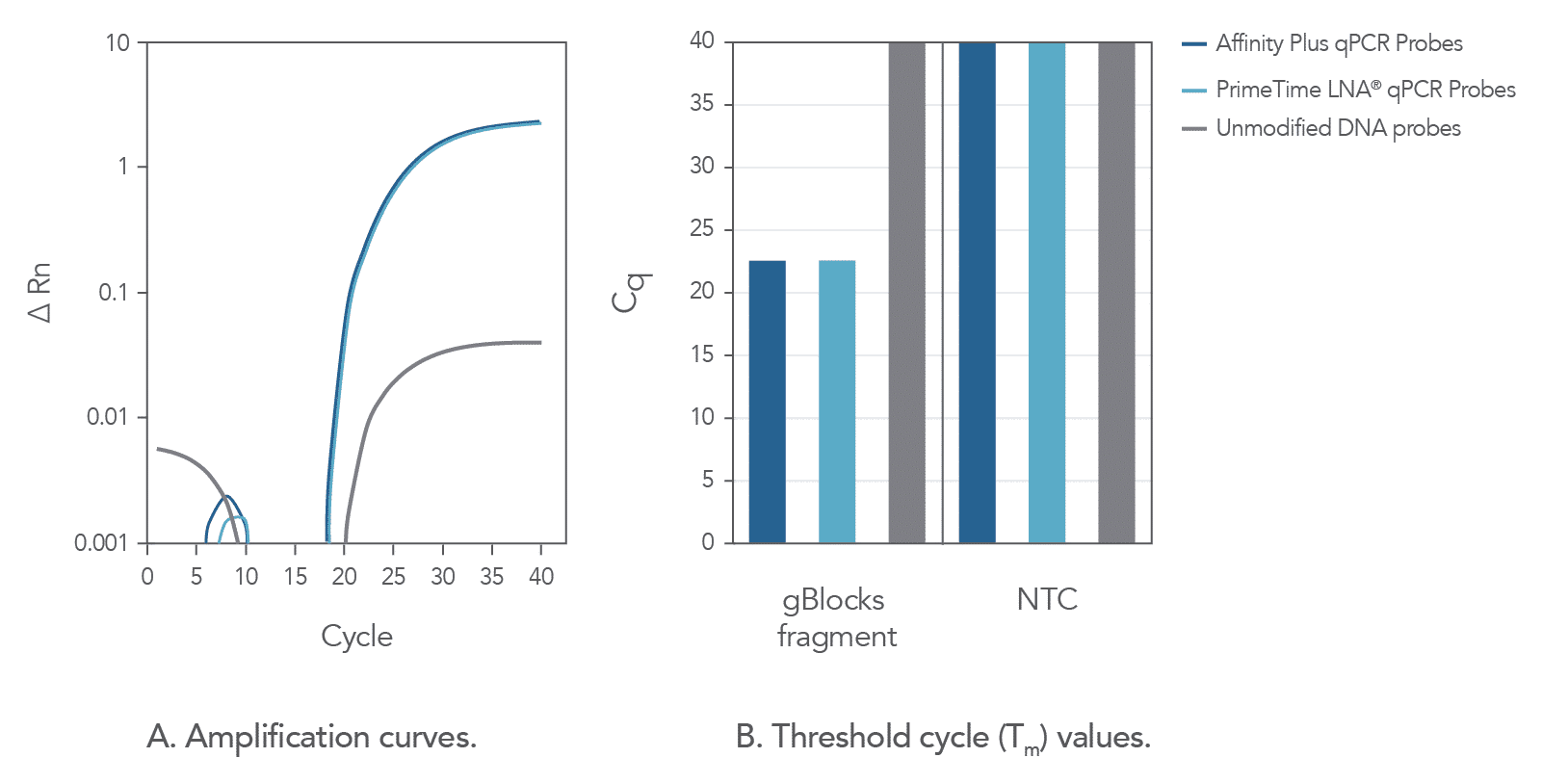
The incorporation of locked nucleic acids into Affinity Plus qPCR Probes imparts heightened structural stability, leading to an increased hybridization melting temperature (Tm), and thus helping them discriminate between thermodynamically similar samples (Figure 2). In comparison to unmodified probe sequences, Affinity Plus qPCR Probes have higher melting temperatures and are more stable in qPCR assays. This increased stability also makes it possible to use shorter probe designs compared to unmodified probes, which can be helpful when targeting regions of a limited size. Figure 2 illustrates that Affinity Plus qPCR Probes provide the same amplification profile as PrimeTime™ LNA® qPCR Probes, with both showing more rapid target amplification compared to unmodified probes (review probe sequences used in these assays in Table 1 of the Product Data section on the Affinity Plus qPCR Probes product page).
Adjustable Tm
The incorporation of locked nucleic acid bases in Affinity Plus Oligos and qPCR Probes enhances hybridization affinity and increases melting temperature (Tm). Unlike other modifications used to enhance oligo Tm (e.g., MGB modifications), the Tm of Affinity Plus DNA & RNA Oligos and Affinity Plus qPCR Probes can be adjusted based on the location and number of modifications added to the oligo. Affinity Plus qPCR Probes can be designed to include up to six locked nucleic acid monomers.
Use Affinity Plus DNA & RNA Oligonucleotides for increased hybridization Tm, stability, and nuclease resistance over standard oligonucleotides. Up to 20 locked nucleic acid monomers can be inserted into a single Affinity Plus Oligonucleotide.
Increased SNP target affinity and stability
Use Affinity Plus qPCR Probes for SNP genotyping, transcript variant identification, and sensitive target identification in challenging samples, such as formalin-fixed, paraffin-embedded (FFPE) tissue or biofluids. The incorporation of locked nucleic acids into Affinity Plus qPCR Probes increases affinity for targeted regions relative to unmodified probes. Thus, it becomes easier to design probes for challenging sequence regions, such as those with high AT content, or where target areas are short. The relatively stronger binding strength also makes it possible to use shorter probes in hybridization assays. Shorter probes can be an asset, especially for rare target identification, such as SNP analysis, as shorter probes have better quenching and a higher signal-to-noise ratio than unmodified longer probes [1].
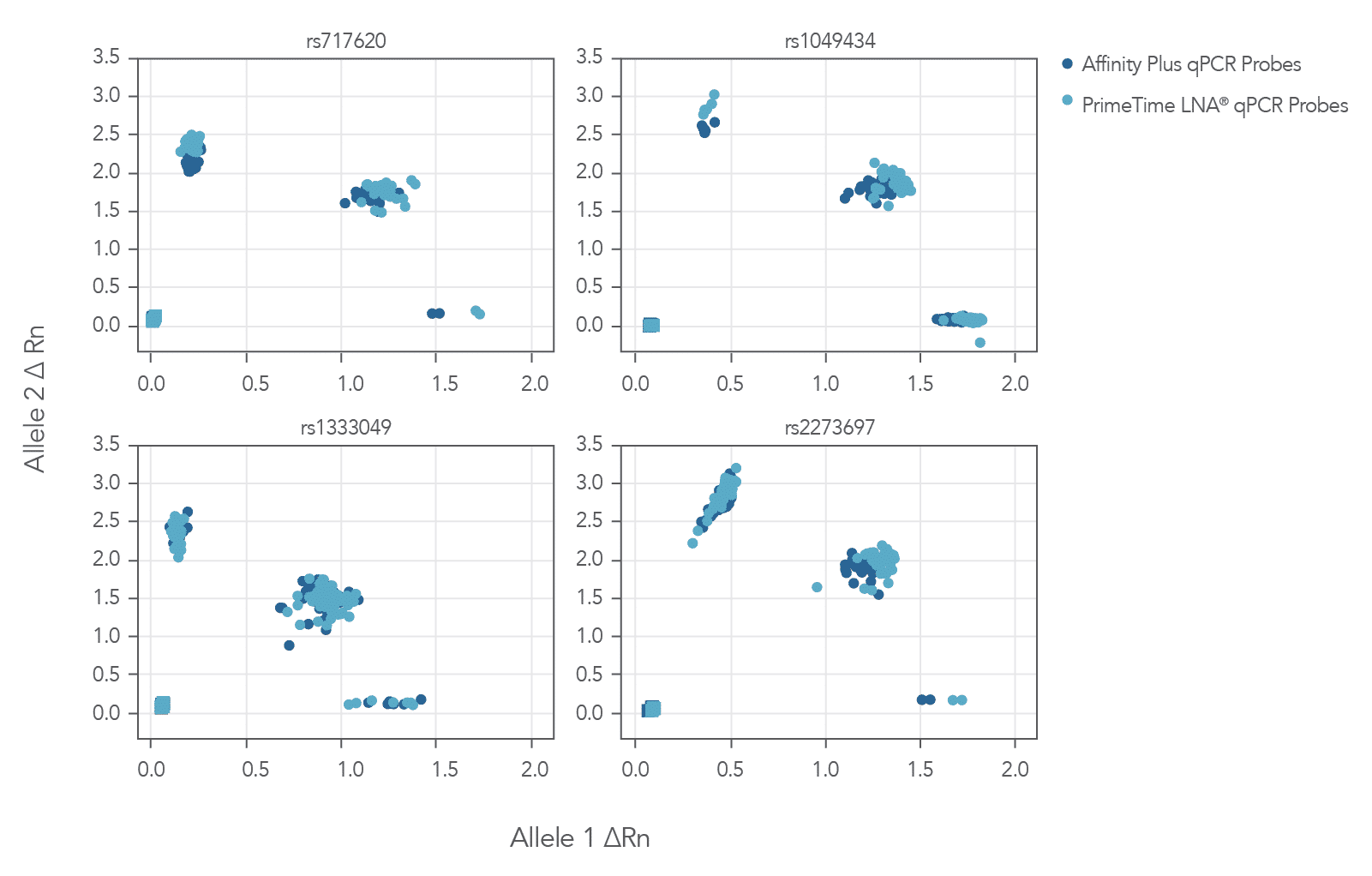
The cluster plots from 4 genotyping assays shown in Figure 3 show that locked nucleic acid-containing Affinity Plus qPCR Probes generate clear, tightly clustered genotyping calls identical to those produced with PrimeTime LNA® Probes. (Review probe sequences used in Table 1 of the Product data section on the Affinity Plus qPCR Probes product page).
In addition, locked nucleic acids improve resistance to exonucleases. Thus, Affinity Plus Oligos and qPCR Probes won’t be degraded as easily.
Broader size and probe quencher options, affordable price
IDT offers Affinity Plus qPCR Probes and Oligonucleotides that are cost-effective and provide identical results to PrimeTime qPCR LNA® Probes, and locked nucleic acid probes and oligos from other vendors.
Learn more or order Affinity Plus DNA & RNA Oligonucleotides.
Learn more or order Affinity Plus qPCR Probes.
*LNA is a registered trademark of Qiagen, Inc.
References
- Ugozzoli LA, Latorra D, Puckett R, et al. Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal Biochem. 2004;324(1):143-152.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. RUO22-1593_001