gBlocks™ & gBlocks HiFi Gene Fragments
gBlocks Gene Fragments and gBlocks HiFi Gene Fragments are double-stranded DNA fragments up to 3000 bp in length, designed for affordable and easy gene construction or modification. Suited for applications such as antibody research and CRISPR-mediated genome editing, qPCR and NGS controls, and more. Drive your projects to completion faster with these high-fidelity fragments.
Ordering
gBlocks Gene Fragments in tubes
NEW Lower pricing on THE original gBlocks!
- A, T, C, and G residues only.
- Delivered dry and normalized to 250, 500, or 1000 ng, depending on length.
- Shipped as soon as 2 business days.
gBlocks Gene Fragments in plates
NEW Lower pricing on THE original gBlocks!
- Resuspended in 25 µL of nuclease-free water (concentration: 10ng/µL).
- Shipped on dry ice as soon as 4 business days of order confirmation (excluding Fridays).
- Orders require a minimum of 24 fragments per plate*.
*Plates with less than 24 fragments will incur an additional charge at checkout.
gBlocks HiFi Gene Fragments in tubes
- There are no order minimums for tubes.
- Shipped dry within 6–10 business days of order confirmation (excluding Fridays).
gBlocks Gene Fragment Libraries in tubes
- Delivered dry, normalized to 200 ng.
- Up to 18 N or K mixed bases.
- Libraries are not available in plate format.
1BD, business days. Shipping time is dependent on length and complexity of the dsDNA fragment(s) ordered. In a few cases, shipping time may exceed the estimated time.
Request a consultation
Working on protein or enzyme engineering? Your time is valuable and we’ll prioritize your inquiry. Click on “Request a consultation” to provide brief information about your project, and we’ll be in touch to discuss it ASAP.
Request a consultationProduct details
gBlocks Gene Fragments
gBlocks Gene Fragments are double-stranded DNA fragments 125–3000 bp in length with a median error rate of less than 1:5000. They are manufactured with the same industry-leading, high-fidelity synthesis chemistries that were developed for our Ultramer™ DNA Oligos.
Each gBlocks Gene Fragment goes through a quality control process, which includes size verification by capillary electrophoresis. This rigorous testing ensures that most recombinant colonies obtained from cloning each gBlocks Gene Fragment will contain the desired insert. More complex sequences may need the end user to sequence additional clones.
gBlocks HiFi Gene Fragments
gBlocks HiFi Gene Fragments are double-stranded DNA fragments between 1000–3000 bp that are sequence-verified via NGS with a median error rate of less than 1:12,000. These high-quality, high-fidelity fragments facilitate the assembly of large and complex sequences, matching both the length and accuracy needed to minimize the introduction of unwanted substitution or deletion errors.
With either gBlocks or gBlocks HiFi Gene Fragments, you can easily assemble and clone your DNA fragment into the vector of your choice using a variety of cloning methods. For added flexibility, you can order gBlocks Gene Fragments with or without a 5′‑phosphate group.
gBlocks Gene Fragment Libraries
gBlocks Gene Fragment Libraries are pooled gBlocks fragments of 251–500 bp in total length. gBlocks Gene Fragment Libraries are ideal for generating recombinant antibodies or for protein engineering, allowing researchers to generate hundreds of thousands of constructs within a reasonable budget. The variable regions can be up to 18 consecutive N (any base) or K (G or T) bases long and must be at least 125 bp from either end of the gene fragment (Figure 1).
Figure 1. Ordering format for gBlocks Gene Fragment Libraries. Placing a K mixed base in the third position of codons eliminates the TAA and TGA stop codons from being included in the gene fragments libraries, leaving only TAG as the possible stop codon.
For gBlocks Gene Fragment Libraries, each constant region is verified similarly to standard gBlocks Gene Fragments. The final library product is size-verified by capillary electrophoresis.
Product data
High fidelity and purity for gene assembly
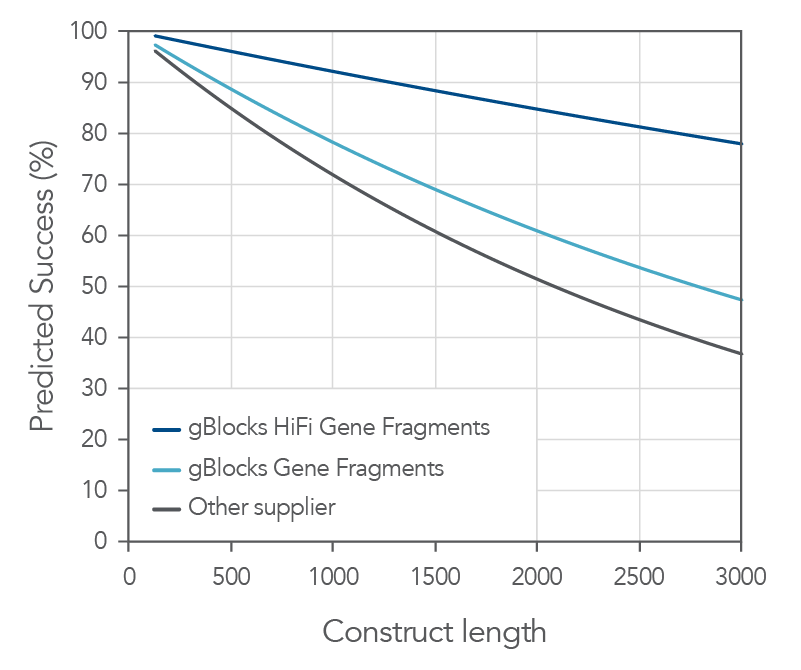
Both gBlocks and gBlocks HiFi Gene Fragments demonstrate consistent high sequence fidelity and purity across various lengths.
Figure 2. IDT gBlocks and gBlocks HiFi Gene Fragments demonstrate high probability of cloning success. With a median error rate of 1:12,000 for gBlocks HiFi Gene Fragments, IDT Gene Fragments demonstrate a high probability of first-time cloning success. Based on data derived from NGS sequencing of gene fragments, gBlocks Gene Fragments and gBlocks HiFi Gene Fragments are up to 45% more likely to give a correct clone the first time when cloning fragments up to 3000 bps compared to an alternate supplier.
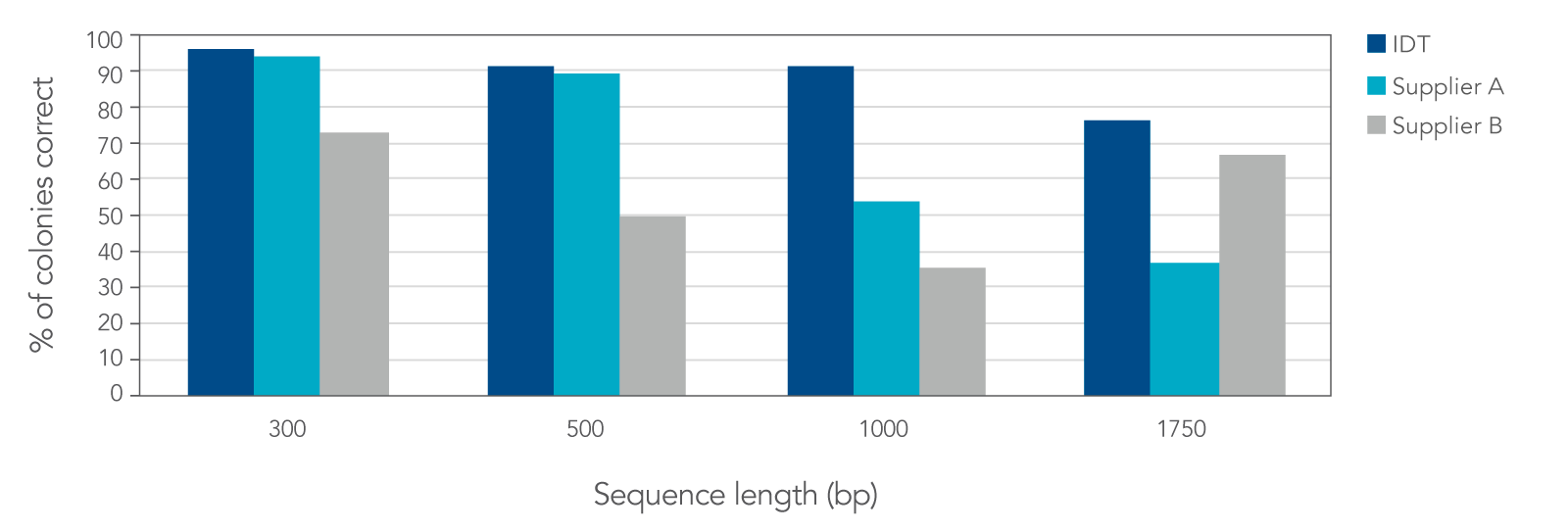
IDT gene fragment data demonstrate improved fidelity versus that from other suppliers
In a head-to-head comparison study against two other DNA synthesis suppliers, IDT’s proprietary synthesis and error correction processes resulted in gene fragments with lower error rates. Low error rates ensure more correct clones, allowing researchers to reduce the number of colonies needed to screen by as much as 50%.
Figure 3. IDT gBlocks and gBlocks HiFi Gene Fragments produce a higher percentage of correct colonies when compared to two other suppliers. Based on screening and sequencing of 24 colonies per sequence, IDT’s fragments were the only fragments to have greater than 75% correct colonies with the desired full-length sequence, and the only gene fragments to achieve greater than 90% correct colonies for fragments that were 1 kb or more in length.
Minimal screening effort needed to find your correct clone
Using IDT gene fragments can reduce the time and expense of screening colonies compared to fragments from other suppliers. Cloning efficiency is affected by many factors, including the cloning method used, the stability of the cell line and plasmid, vector preparation, and toxicity or stress from expression of coding sequences. The values in Table 1 represent typical screening numbers needed when using a seamless cloning method.
Table 1. gBlocks and gBlocks HiFi Gene Fragments reduce the time and expense of screening colonies. The approximate number of colonies to screen for a 90% chance of getting a correct clone.
| Length (bp) | gBlocks Gene Fragments | gBlocks HiFi Gene Fragments |
|---|---|---|
| ≤1000 | 2–3 | N/A |
| 1001–2000 | 3 | 2 |
| 2001–3000 | 4 | 2 |
Compatible with multiple cloning methods and workflows
IDT gene fragments are compatible with many cloning and assembly kits and automation platforms, allowing easy assembly of your desired construct sequence into your preferred cloning method. Methods include traditional cloning, Gibson Assembly® (New England Biolabs), Golden Gate, Gateway® (Invitrogen), TOPO™/TA cloning (Thermo Fisher), blunt-end cloning, and others.
Resources
Frequently asked questions
I found a mutation in my clone. Is there a problem with the gBlocks™ Gene Fragment?
Fortunately, the large majority of sequences should be correct, so by sequencing a few more colonies you should find a correct clone.
What can I do if I do not get any colonies during my cloning reaction using gBlocks™ Gene Fragments?
The most common source of this issue is an incomplete resuspension of your gBlocks Gene Fragment. The pellets can stick to the side or cap of the tube, and unless you vortex vigorously and incubate the tube at 50°C for 20 minutes, you might not have the DNA concentration you expect. We recommend the following:
- Vortex your tube, and incubate at 50°C for 20 minutes.
- Vortex vigorously again and spin the tube down.
- Recheck the concentration of your solution. (In general, it is always a good idea to recheck your concentration after you resuspend a DNA sample.)
What should I do if the IDT online ordering tool says that my sequence is too complex to order, but I need it as is?
In most cases, our system has found a region with either very high G/C content or several small repeats that would prevent us from synthesizing your sequence.
If your sequence contains a coding region, you can use codon optimization to reduce the complexity. Codon optimization uses synonymous codons to retain the final amino acid sequence that is expressed while generating a sequence that we can synthesize.
If your sequence is not a coding region, we might be able to synthesize the sequence as a custom gene, which is delivered in a basic pUC vector, rather than just a linear, dsDNA fragment. For assistance, send your sequences and any questions to:
amr-applicationsupport@idtdna.com (for customers in the United States or Canada)
emea-applicationsupport@idtdna.com (for customers in Europe)
apac-applicationsupport@idtdna.com (for customers in Asia Pacific, Australia, New Zealand, Japan, and South Korea)
How can I use PCR to add overlapping regions to my gBlocks™ Gene Fragments that I will use in a seamless cloning reaction?
We do not recommend using PCR to add the overhangs, because PCR can preferentially amplify smaller byproducts in the prep, leading to issues with the cloning reaction and increasing the number of colonies you need to screen to find the correct full-length insert. Instead, build the overhangs into the gBlocks Gene Fragment itself. The amount of material you will receive is enough for a couple of assembly reactions.
The additional PCR step is not required to generate additional material and adds an additional enzymatic step to the protocol that can introduce errors.
What are optimum storage conditions for a gBlocks™ HiFi Gene Fragment?
IDT ships gBlocks HiFi Gene Fragments (IDT) dry unless otherwise requested. While dry, they should be stable almost indefinitely.
After resuspending in a high-quality, molecular-grade water or buffer, pH 7.5–8.0, you should store the DNA at –20°C and if necessary, aliquot to avoid more than 2 or 3 freeze-thaw cycles. Adding a carrier to your resuspension and dilution buffers can help prevent any potential decrease in product concentration.
Should I amplify my gBlocks™ HiFi Gene Fragments when I receive them?
gBlocks HiFi Gene Fragments are clonal products and can be amplified if necessary. However, most applications such as restriction-digest, blunt-end, and Gibson cloning can be performed with the material provided by IDT, and PCR amplification should not be necessary.
If the purpose of the amplification is to add flanking bases to complete a cloning reaction, we strongly recommend ordering the gBlocks HiFi Gene Fragment (IDT) with the required bases rather than adding them afterwards using PCR.
If you choose to amplify your gBlocks HiFi Gene Fragment, use the minimum amount of starting template required by your PCR kit protocol, limit the cycle number to 10–12, and use a high-fidelity polymerase.
How do I design my gBlocks™ HiFi Gene Fragment for restriction cloning?
Cloning of gBlocks HiFi Gene Fragments is similar to cloning a very pure PCR product. For restriction cloning, it is important to add 6–8 nt at the ends of the fragment, beyond the restriction recognition sequence.
Most restriction enzymes will not cut efficiently, or at all, if the fragment terminates precisely at the restriction site.
Can I get gBlocks™ HiFi Gene Fragments outside of the 1000–3000 bp range?
We currently only offer gBlocks™ HiFi Gene Fragments between 1000–3000 bp.
How is a gBlocks™ HiFi Gene Fragment different from a gBlocks Gene Fragment?
The combination of speed, cost, and purity make gBlocks™ Gene Fragments an ideal solution for most applications, but in some applications, such as the assembly of larger constructs, starting with a fragment with a lower error rate may significantly reduce costs.
gBlocks HiFi Gene Fragments use a proprietary manufacturing technique that can generate double-stranded DNA fragments with average error rates of 1:12,000, which is approximately 2.5–4x fewer errors than other commercially available gene fragments.
What is the length of gBlocks™ Gene Fragments that IDT can synthesize?
IDT can synthesize gBlocks Gene Fragments between 125 bp and 3 kb in length.
However, if you require a double-stranded DNA construct outside of this size range, we offer 2 options:
- We may be able to accept the sequence for synthesis as a custom gene.
- Our support scientists can assist in splitting larger sequences into multiple gBlocks Gene Fragments that can be used in an assembly reaction to generate the full-length construct.
If you are interested in a custom gene quote or assistance in splitting your sequence into sub-fragments, please email your sequences to:
amr-applicationsupport@idtdna.com (for customers in the United States or Canada)
emea-applicationsupport@idtdna.com (for customers in Europe)
apac-applicationsupport@idtdna.com (for customers in Asia Pacific, Australia, New Zealand, Japan, and South Korea)
Should I amplify my gBlocks™ Gene Fragments when I receive them?
We do not recommend amplifying gBlocks Gene Fragments >1 kb in length.
gBlocks Gene Fragments are synthesized from several smaller overlapping fragments. The final prep may contain some of these smaller byproducts. During PCR, smaller products are more efficiently amplified than longer ones, and at the exponential phase of PCR, smaller products in the reaction will overwhelm the generation of the full-length correct product. When run on the gel, these amplification products appear as a smear or smaller bands than expected.
Most applications such as restriction digest, blunt end, and seamless cloning can be performed with the material provided by IDT, and PCR amplification should not be necessary. If the purpose of the amplification is to add flanking bases to complete a cloning reaction, we strongly recommend ordering the gBlocks Gene Fragment with the required bases rather than adding them afterwards using PCR.
If you must amplify your gBlocks Gene Fragment, use the minimum amount of starting template required by your PCR kit protocol, limit the cycle number to 10–12, and use a high-fidelity polymerase.
How long does it take to make a gBlocks™ Gene Fragment?
Synthesis time depends on the length of the gBlocks fragment.
| Length of gBlocks Gene Fragment | Time to ship after order confirmation (business days) |
|---|---|
| Up to 750 bp | 2–4 |
| 751–1000 bp | 3–5 |
| More than 1000 bp | 5–8 |
What is the difference between a gBlocks™ Gene Fragment and an eBlocks™ Gene Fragment?
Here is a comparison of gBlocks and eBlocks Gene Fragments:
| gBlocks Gene Fragments | eBlocks Gene Fragments | |
|---|---|---|
| Double-stranded DNA | ✓ | ✓ |
| Stringent QC process • Median error rate • Synthesis & assembly |
1:5000 May rework to meet QC standards |
1:5000 No rework† |
| Format | Plates or tubes | Plate |
| Length | 125–3000 bp | 300–1500 bp |
† Any eBlocks Gene Fragment that does not meet QC standards after the first attempt will be cancelled and refunded.
What are optimum storage conditions for a gBlocks™ Gene Fragment?
IDT ships gBlocks Gene Fragments dry unless otherwise requested. While dry, they should be stable up to two years.
After resuspending in a high-quality, molecular-grade water or buffer, pH 7.5–8.0, you should store the DNA at –20°C and if necessary, aliquot to avoid more than 2 or 3 freeze-thaw cycles.
For gBlocks Gene Fragments at concentrations lower than 1 ng/µL, we recommend resuspending in water or buffer containing 0.1–1.0 mg/mL tRNA (Sigma) or poly A (Sigma). Use this same buffer + carrier solution for subsequent dilutions. Without the carrier, we have observed a steady decrease in measured concentrations of these dilute solutions, even in low-bind tubes. This may be caused by the very high purity of the gBlocks Gene Fragment prep. Because gBlocks Gene fragments are completely synthetic, they lack the normal cellular debris found in DNA isolated from natural sources.
Without these natural carriers, the gBlocks Gene Fragments bind irreversibly to the tube plastic over time, leading to the observed decrease in concentration. Adding a carrier to your resuspension and dilution buffers can help prevent this loss of product.
Why am I limited to 18 N or K mixed bases in my gBlocks™ Gene Fragment?
We decided to limit variable regions to 18 mixed bases to give customers the best overall pool of library constructs. Since most functional screens cover 1,000–100,000 recombinant colonies, allowing 18 mixed bases should be adequate for producing meaningful results.
What types of sequence motifs should be avoided when ordering gBlocks™ Gene Fragments?
What quality checks do gBlocks™ Gene Fragments with variable bases receive?
The constant regions are gBlocks Gene Fragments and undergo size verification by capillary electrophoresis.
The variable regions cannot currently receive complete analysis (using NGS, which would be needed for this analysis, would be cost prohibitive). We rely on a validated process that we have experimentally confirmed by NGS that ensures >80% of DNA species are present in the final material shipped.
What if I need a gBlocks™ Gene Fragment library that is more complex than what you offer online?
We are working hard at offering more complex libraries in the future.
Help us prioritize by sharing what you need from us in as much detail as possible (i.e., show us sequences including mixed bases, annotations, drawings, etc.).
You can send your requests to:
amr-applicationsupport@idtdna.com (for customers in the United States or Canada)
emea-applicationsupport@idtdna.com (for customers in Europe)
apac-applicationsupport@idtdna.com (for customers in Asia Pacific, Australia, New Zealand, Japan, and South Korea)
What are the N and K mixed bases that can be incorporated into the variable regions of gBlocks™ Gene Fragments?
Additionally, using NNK codons removes 2 out of 3 possible stop codons, which also limits the number of sequences in a library that produce unwanted truncated gene products.
Should I order my gBlocks™ Gene Fragment with the 5′ phosphate modification?
The 5′ phosphate is required for traditional blunt-end cloning. Do not add 5′ phosphate for Zero Blunt TOPO® cloning (check manufacturer information).
Biosecurity
Sequence Information is secure and confidential at IDT. Please see our Confidentiality Statement for more information. All online ordering steps, including sequence entry and your choice of parameters, are also secure and protected.
We screen the sequence of every gene, and gene fragment order we receive to (1) identify any regulated and other potentially dangerous pathogen sequences, and (2) verify that IDT’s gene customers are legitimate scientists engaged in beneficial research.
IDT is among the five founding members of the International Gene Synthesis Consortium (IGSC) and helped to create the IGSC’s Harmonized Screening Protocol. The Harmonized Screening Protocol describes the gene sequence and customer screening practices that IGSC member companies employ to prevent the misuse of synthetic genes. IDT takes the steps set out in the Harmonized Screening Protocol to screen the sequences of ordered genes and the prospective customers who submit those orders.
For more information about the IGSC and the Harmonized Screening Protocol, please visit the website at www.genesynthesisconsortium.org.

In October 2010, the United States government issued final Screening Framework Guidance for Providers of Synthetic Double-Stranded DNA, describing how commercial providers of synthetic genes should perform gene sequence and customer screening. IDT and the other IGSC member companies supported the adoption of the Screening Framework Guidance, and IDT follows that Guidance in its application of the Harmonized Screening Protocol. For more information, please see 75 FR 62820 (Oct. 13, 2010), or https://federalregister.gov/a/2010-25728.