Frequently asked questions
Our team has assembled a list of frequently asked questions to help you find answers quickly. Filter using one or more categories to focus on specific topics, or use the search bar to perform a text search.
Search all FAQs:
Can I use a CRISPR-Cas9 crRNA protospacer sequence that is shorter than 20 nt?
The Alt-R™ CRISPR-Cas9 crRNA ordering tool (accessible here) accommodates 19 and 20 nucleotide (nt) protospacer sequences; however, we recommend 20 nt sequences for most experiments. Other formats can be ordered as custom RNAs.
There are reports in the literature suggesting that CRISPR-Cas9 nuclease specificity can be improved through use of truncated guide RNAs [1]. For example, 17 nt protospacer elements have been reported to reduce off-target effects.
In contrast, our research investigating the effect of shorter protospacer element length on CRISPR-Cas9 nuclease specificity demonstrated that 20 nt protospacer elements were optimal, with 19 nt protospacers providing similar strong editing efficacy in most cases. When using Alt-R S.p. HiFi Cas9 Nuclease, 20 nt protospacer sequences provide the greatest amount of genomic editing.
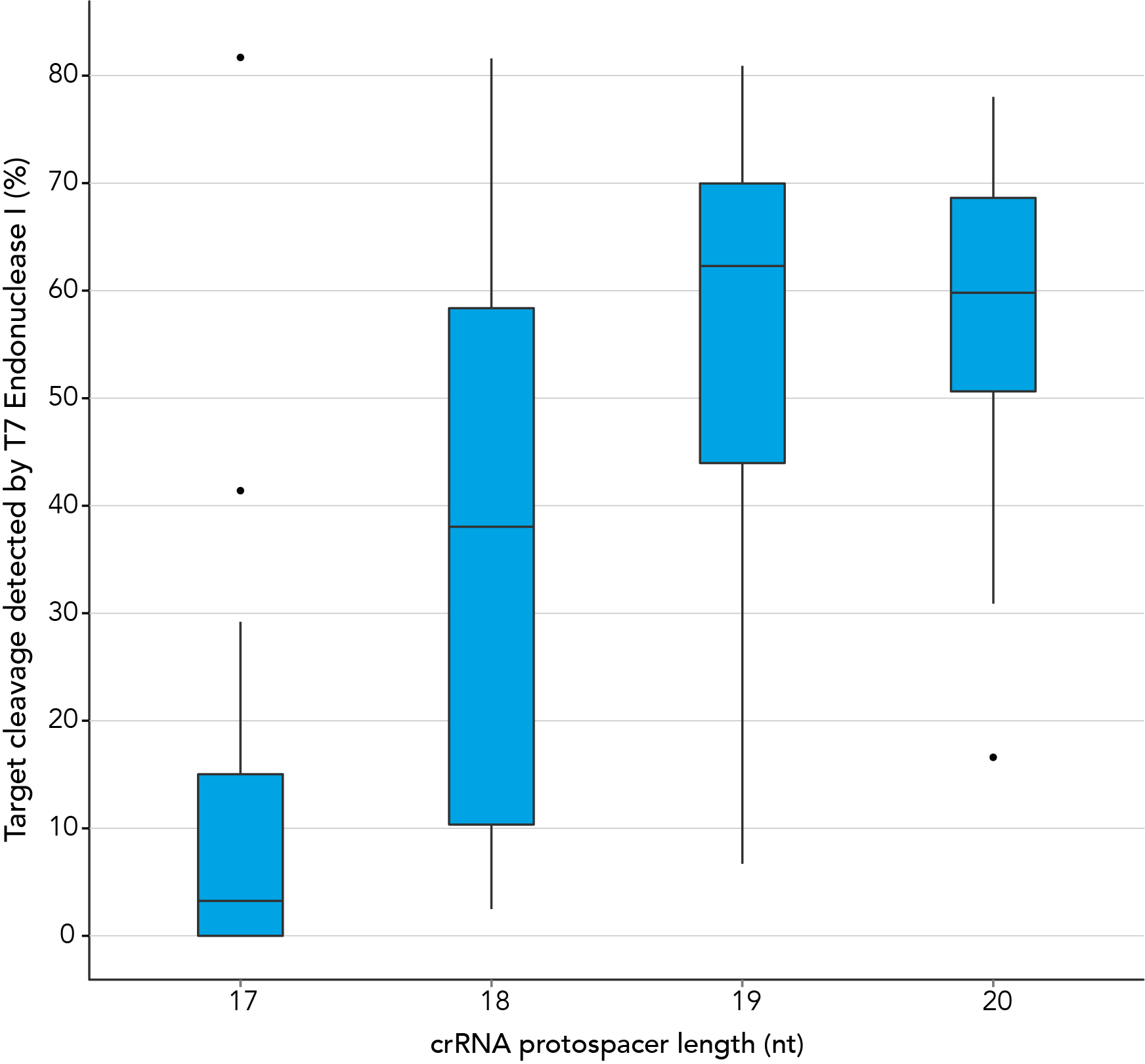 Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.
Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279-284.