How to convert from nanograms to copy number
Researchers performing qPCR will often create a standard curve based on nanograms of amplicon, and then need to convert the resulting nanograms detected to copy number. The formula for making this conversion is:
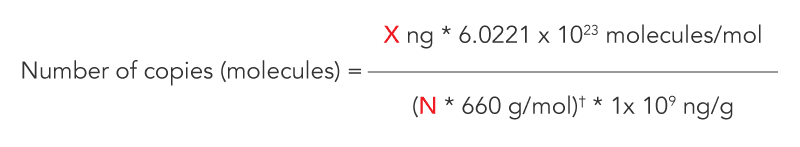
Where: X = amount of amplicon (ng) N = length of dsDNA amplicon 660 g/mol = average mass of 1 bp dsDNA 6.022 x 1023 = Avogadro’s constant 1 x 109 = Conversion factor
† The actual oligonucleotide MW (in Daltons or g/mol) is provided on the IDT Spec Sheet for each oligonucleotide and can be substituted for this arithmetic phrase, which only provides an average MW for a sequence of this length.
It is important to note if using a single stranded DNA (ssDNA) oligo as a template, the molecular weight provided on the IDT Spec Sheet can replace the (N * 660 g/mol) factor in the formula in Figure 1. However, if you want to calculate that factor using the length of a ssDNA molecule, then the average mass would be 330 g/mol, instead of 660 g/mol.
Alternatively, you can create the standard curve based on copy number; e.g., ranging from 102–107 copies, in 10-fold increments. Then you can determine copy number directly from the standard curve.
Let a free copy number calculator do it for you!
A convenient, free-access copy number calculator to convert nanograms to copy number for real time PCR (RT-PCR) was developed by former IDT customer, Andrew Staroscik, who has since left the bench.
The copy number calculator can be found here.
RUO23-1753_001.1